Deconvolution: Software Confocal
Deconvolution: Software Confocal
Widefield microscopy resolution can approach the diffraction limit in the X and Y optical planes but suffers from lower resolution in the Z axis. Confocal microscopy was introduced by Marvin Minsky in 1953 to increase resolution, especially in the z-axis, by using a pinhole to eliminate out of focus light contribution. This allows thin Z-slices to be acquired and reconstructed into new 2D and 3D views. This capability comes at the cost of time and phototoxicity, not to mention the cost of the hardware itself.
“Widefield microscopy uses low power spread over every pixel for the full duration of the image, but laser scanning confocal microscopy focuses all of the light onto a single pixel for a short period of time and then raster scans the pixels. Using identical total power for widefield and confocal imaging, the instantaneous power (i.e., intensity) is much higher in laser scanning. For a one second 512×512 image, the laser scanning intensity is 250,000-fold larger than in widefield illumination.“ Piston et al, Nature Methods, 13:557–562 (2016).
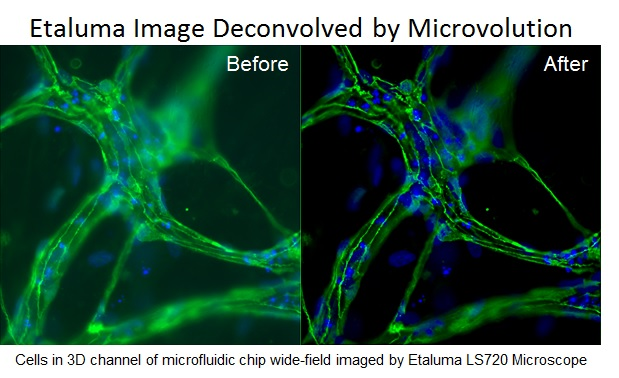
As live cell imaging becomes more common, especially with 3D models, phototoxicity, cost, and complexity of the instruments must be lowered. Like many things in technology, a software solution often supersedes early hardware solutions.
Deconvolution of images through the use of a point spread function (PSF) was first described in the early 1970s by the namesakes of the Richardson-Lucy maximum likelihood algorithm (Richardson, 1972, J. Opt. Soc. Am 62:55-59, Lucy, 1974, Astron. J. 79:745. Since then, its use in widefield microscopy has generated confocal-like images from widefield optics. These algorithms are intensive and iterative and can take minutes to hours to operate on a stack of images. Commercial deconvolution software is expensive and runs on restricted operating system platforms, which may not be suitable for resource limited users.
Microvolution LLC (Sunnyvale, CA) has developed a parallelized software method that deconvolves 3D images over 100 times faster than conventional approaches by utilizing graphics processor units (GPU)s developed for gaming. Etaluma has partnered with Microvolution and now offers this capability in our line of LS Microscopes.
The largest speed gains in the Microvolution software are accomplished by implementing the convolution steps of the R-L algorithm on the GPU, noting that a convolution in real space is equivalent to a pointwise multiplication in Fourier space. The CUFFT library provided by NVIDIA is a highly tuned FFT implementation that is generalized to arbitrary dimensions, allowing the software to operate efficiently on images of any size. Processing all arithmetic operations in parallel on the GPU further accelerates software execution, resulting in the ability to deconvolve real-world 3D microscopy images in a fraction of a second. The software deconvolves images ~10 times faster than they can be collected, and about as fast as they can be written to the hard drive. This speed allows the possibility of displaying the deconvolved images by the microscope control software as fast as the data are collected. By showing deconvolved images immediately, the user can adjust experimental parameters on the fly. The software even automatically deconvolves each color of multi-color images.
The advantages of widefield fluorescence microscopy in concert with deconvolution, what we refer to as “Software Confocal”, over traditional laser scanning confocal microscopy are sensitivity, lack of photo-damage, lower cost, smaller size, and simplicity. The use of Software Confocal also enables multi-sample 3D imaging of living specimens in microplates and dishes. We envision Microvolution will broadly increase the information content and improve the accuracy of 3D fluorescence microscopy across many applications in cell biology.
Copyright © 2018 Etaluma, Inc. All rights reserved. Website design by Eskisse
